Kriging¶
Kriging is an interpolating model that is a linear combination of a known function \(f_i({\bf x})\) which is added to a realization of a stochastic process \(Z({\bf x})\)
\(Z({\bf x})\) is a realization of a stochastic process with mean zero and spatial covariance function given by
where \(\sigma^2\) is the process variance, and \(R\) is the correlation. Four types of correlation functions are available in SMT.
Exponential correlation function (Ornstein-Uhlenbeck process):
Squared Exponential (Gaussian) correlation function:
Exponential correlation function with a variable power:
Matérn 5/2 correlation function:
Matérn 3/2 correlation function:
Exponential Squared Sine correlation function:
These correlation functions are called by ‘abs_exp’ (exponential), ‘squar_exp’ (Gaussian), ‘matern52’,’matern32’ and ‘squar_sin_exp’ in SMT.
The deterministic term \(\sum\limits_{i=1}^k\beta_i f_i({\bf x})\) can be replaced by a constant, a linear model, or a quadratic model. These three types are available in SMT.
In the implementations, data are normalized by substracting the mean from each variable (indexed by columns in X), and then dividing the values of each variable by its standard deviation:
More details about the Kriging approach could be found in [1].
Kriging with categorical or integer variables¶
The goal is to be able to build a model for mixed typed variables. This algorithm has been presented by Garrido-Merchán and Hernández-Lobato in 2020 [2].
To incorporate integer (with order relation) and categorical variables (with no order), we used continuous relaxation. For integer, we add a continuous dimension with the same bounds and then we round in the prediction to the closer integer. For categorical, we add as many continuous dimensions with bounds [0,1] as possible output values for the variable and then we round in the prediction to the output dimension giving the greatest continuous prediction.
A special case is the use of the Gower distance to handle mixed integer variables (hence the gower kernel/correlation model option). See the MixedInteger Tutorial for such usage.
More details available in [2]. See also Mixed integer surrogate.
Implementation Note: Mixed variables handling is available for all Kriging models (KRG, KPLS or KPLSK) but cannot be used with derivatives computation.
Usage¶
Example 1¶
import numpy as np
import matplotlib.pyplot as plt
from smt.surrogate_models import KRG
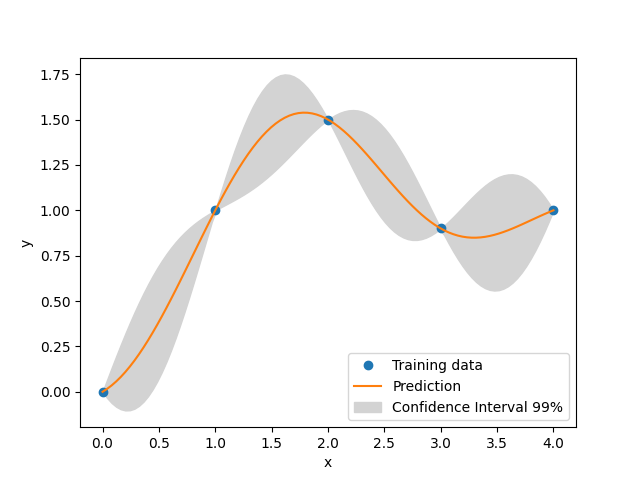
xt = np.array([0.0, 1.0, 2.0, 3.0, 4.0])
yt = np.array([0.0, 1.0, 1.5, 0.9, 1.0])
sm = KRG(theta0=[1e-2])
sm.set_training_values(xt, yt)
sm.train()
num = 100
x = np.linspace(0.0, 4.0, num)
y = sm.predict_values(x)
# estimated variance
s2 = sm.predict_variances(x)
# derivative according to the first variable
_dydx = sm.predict_derivatives(xt, 0)
_, axs = plt.subplots(1)
# add a plot with variance
axs.plot(xt, yt, "o")
axs.plot(x, y)
axs.fill_between(
np.ravel(x),
np.ravel(y - 3 * np.sqrt(s2)),
np.ravel(y + 3 * np.sqrt(s2)),
color="lightgrey",
)
axs.set_xlabel("x")
axs.set_ylabel("y")
axs.legend(
["Training data", "Prediction", "Confidence Interval 99%"],
loc="lower right",
)
plt.show()
___________________________________________________________________________
Kriging
___________________________________________________________________________
Problem size
# training points. : 5
___________________________________________________________________________
Training
Training ...
Training - done. Time (sec): 0.1566455
___________________________________________________________________________
Evaluation
# eval points. : 100
Predicting ...
Predicting - done. Time (sec): 0.0029492
Prediction time/pt. (sec) : 0.0000295
___________________________________________________________________________
Evaluation
# eval points. : 5
Predicting ...
Predicting - done. Time (sec): 0.0005305
Prediction time/pt. (sec) : 0.0001061
Example 2 with mixed variables¶
import numpy as np
import matplotlib.pyplot as plt
from smt.surrogate_models import KRG
from smt.applications.mixed_integer import MixedIntegerKrigingModel
from smt.utils.design_space import DesignSpace, IntegerVariable
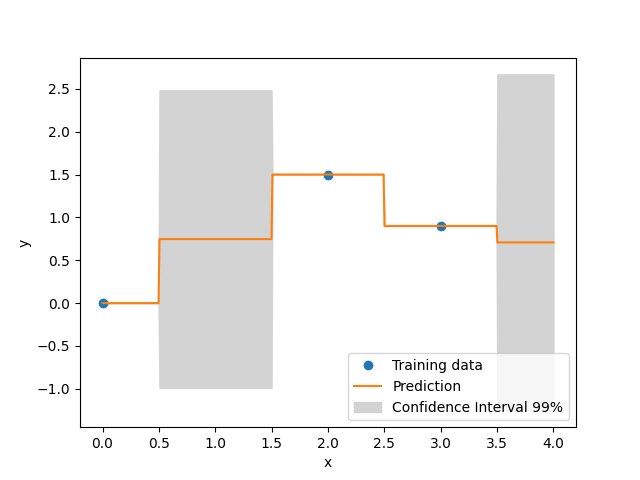
xt = np.array([0.0, 2.0, 3.0])
yt = np.array([0.0, 1.5, 0.9])
design_space = DesignSpace(
[
IntegerVariable(0, 4),
]
)
sm = MixedIntegerKrigingModel(
surrogate=KRG(design_space=design_space, theta0=[1e-2], hyper_opt="Cobyla")
)
sm.set_training_values(xt, yt)
sm.train()
num = 500
x = np.linspace(0.0, 4.0, num)
y = sm.predict_values(x)
# estimated variance
s2 = sm.predict_variances(x)
fig, axs = plt.subplots(1)
axs.plot(xt, yt, "o")
axs.plot(x, y)
axs.fill_between(
np.ravel(x),
np.ravel(y - 3 * np.sqrt(s2)),
np.ravel(y + 3 * np.sqrt(s2)),
color="lightgrey",
)
axs.set_xlabel("x")
axs.set_ylabel("y")
axs.legend(
["Training data", "Prediction", "Confidence Interval 99%"],
loc="lower right",
)
plt.show()
___________________________________________________________________________
Evaluation
# eval points. : 500
Predicting ...
Predicting - done. Time (sec): 0.0096734
Prediction time/pt. (sec) : 0.0000193
Example 3 with noisy data¶
import numpy as np
import matplotlib.pyplot as plt
from smt.surrogate_models import KRG
# defining the toy example
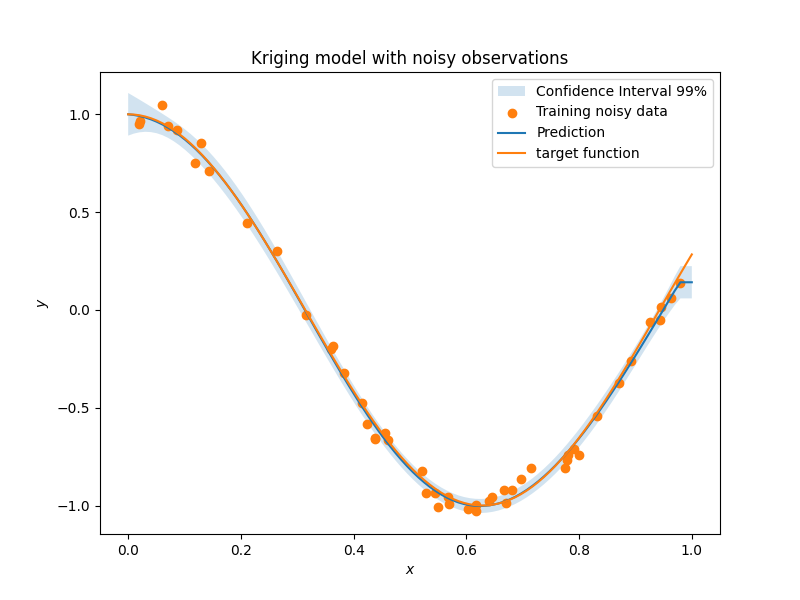
def target_fun(x):
import numpy as np
return np.cos(5 * x)
nobs = 50 # number of obsertvations
np.random.seed(0) # a seed for reproducibility
xt = np.random.uniform(size=nobs) # design points
# adding a random noise to observations
yt = target_fun(xt) + np.random.normal(scale=0.05, size=nobs)
# training the model with the option eval_noise= True
sm = KRG(eval_noise=True)
sm.set_training_values(xt, yt)
sm.train()
# predictions
x = np.linspace(0, 1, 100).reshape(-1, 1)
y = sm.predict_values(x) # predictive mean
var = sm.predict_variances(x) # predictive variance
# plotting predictions +- 3 std confidence intervals
plt.rcParams["figure.figsize"] = [8, 4]
plt.fill_between(
np.ravel(x),
np.ravel(y - 3 * np.sqrt(var)),
np.ravel(y + 3 * np.sqrt(var)),
alpha=0.2,
label="Confidence Interval 99%",
)
plt.scatter(xt, yt, label="Training noisy data")
plt.plot(x, y, label="Prediction")
plt.plot(x, target_fun(x), label="target function")
plt.title("Kriging model with noisy observations")
plt.legend(loc=0)
plt.xlabel(r"$x$")
plt.ylabel(r"$y$")
___________________________________________________________________________
Kriging
___________________________________________________________________________
Problem size
# training points. : 50
___________________________________________________________________________
Training
Training ...
Training - done. Time (sec): 0.3628881
___________________________________________________________________________
Evaluation
# eval points. : 100
Predicting ...
Predicting - done. Time (sec): 0.0112629
Prediction time/pt. (sec) : 0.0001126
Options¶
Option |
Default |
Acceptable values |
Acceptable types |
Description |
|---|---|---|---|---|
print_global |
True |
None |
[‘bool’] |
Global print toggle. If False, all printing is suppressed |
print_training |
True |
None |
[‘bool’] |
Whether to print training information |
print_prediction |
True |
None |
[‘bool’] |
Whether to print prediction information |
print_problem |
True |
None |
[‘bool’] |
Whether to print problem information |
print_solver |
True |
None |
[‘bool’] |
Whether to print solver information |
poly |
constant |
[‘constant’, ‘linear’, ‘quadratic’] |
[‘str’] |
Regression function type |
corr |
squar_exp |
[‘pow_exp’, ‘abs_exp’, ‘squar_exp’, ‘squar_sin_exp’, ‘matern52’, ‘matern32’] |
[‘str’] |
Correlation function type |
pow_exp_power |
1.9 |
None |
[‘float’] |
Power for the pow_exp kernel function (valid values in (0.0, 2.0]). This option is set automatically when corr option is squar, abs, or matern. |
categorical_kernel |
MixIntKernelType.CONT_RELAX |
[<MixIntKernelType.CONT_RELAX: ‘CONT_RELAX’>, <MixIntKernelType.GOWER: ‘GOWER’>, <MixIntKernelType.EXP_HOMO_HSPHERE: ‘EXP_HOMO_HSPHERE’>, <MixIntKernelType.HOMO_HSPHERE: ‘HOMO_HSPHERE’>, <MixIntKernelType.COMPOUND_SYMMETRY: ‘COMPOUND_SYMMETRY’>] |
None |
The kernel to use for categorical inputs. Only for non continuous Kriging |
hierarchical_kernel |
MixHrcKernelType.ALG_KERNEL |
[<MixHrcKernelType.ALG_KERNEL: ‘ALG_KERNEL’>, <MixHrcKernelType.ARC_KERNEL: ‘ARC_KERNEL’>] |
None |
The kernel to use for mixed hierarchical inputs. Only for non continuous Kriging |
nugget |
2.220446049250313e-14 |
None |
[‘float’] |
a jitter for numerical stability |
theta0 |
[0.01] |
None |
[‘list’, ‘ndarray’] |
Initial hyperparameters |
theta_bounds |
[1e-06, 20.0] |
None |
[‘list’, ‘ndarray’] |
bounds for hyperparameters |
hyper_opt |
TNC |
[‘Cobyla’, ‘TNC’] |
[‘str’] |
Optimiser for hyperparameters optimisation |
eval_noise |
False |
[True, False] |
[‘bool’] |
noise evaluation flag |
noise0 |
[0.0] |
None |
[‘list’, ‘ndarray’] |
Initial noise hyperparameters |
noise_bounds |
[2.220446049250313e-14, 10000000000.0] |
None |
[‘list’, ‘ndarray’] |
bounds for noise hyperparameters |
use_het_noise |
False |
[True, False] |
[‘bool’] |
heteroscedastic noise evaluation flag |
n_start |
10 |
None |
[‘int’] |
number of optimizer runs (multistart method) |
xlimits |
None |
None |
[‘list’, ‘ndarray’] |
definition of a design space of float (continuous) variables: array-like of size nx x 2 (lower, upper bounds) |
design_space |
None |
None |
[‘BaseDesignSpace’, ‘list’, ‘ndarray’] |
definition of the (hierarchical) design space: use smt.utils.design_space.DesignSpace as the main API. Also accepts list of float variable bounds |
random_state |
41 |
None |
[‘NoneType’, ‘int’, ‘RandomState’] |
Numpy RandomState object or seed number which controls random draws for internal optim (set by default to get reproductibility) |