Surrogate modeling methods¶
SMT contains the surrogate modeling methods listed below.
Usage¶
import numpy as np
import matplotlib.pyplot as plt
from smt.surrogate_models import RBF
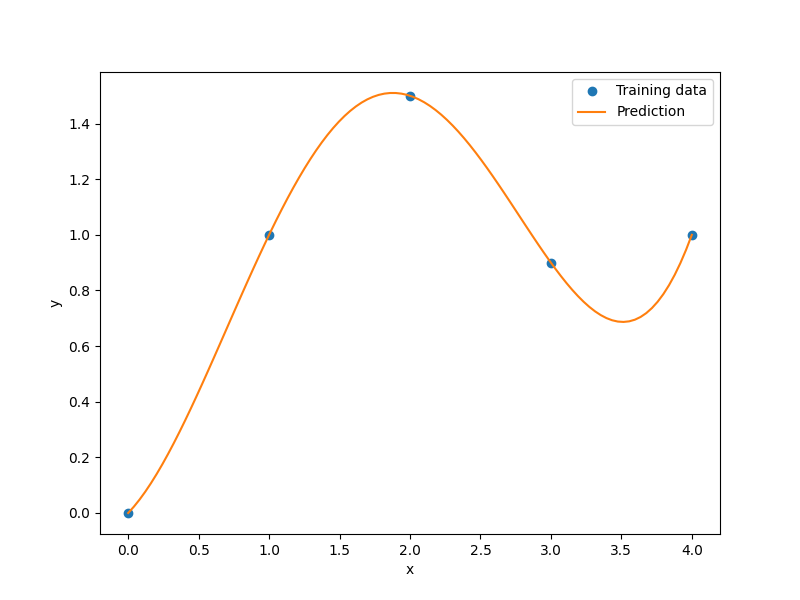
xt = np.array([0.0, 1.0, 2.0, 3.0, 4.0])
yt = np.array([0.0, 1.0, 1.5, 0.9, 1.0])
sm = RBF(d0=5)
sm.set_training_values(xt, yt)
sm.train()
num = 100
x = np.linspace(0.0, 4.0, num)
y = sm.predict_values(x)
plt.plot(xt, yt, "o")
plt.plot(x, y)
plt.xlabel("x")
plt.ylabel("y")
plt.legend(["Training data", "Prediction"])
plt.show()
___________________________________________________________________________
RBF
___________________________________________________________________________
Problem size
# training points. : 5
___________________________________________________________________________
Training
Training ...
Initializing linear solver ...
Performing LU fact. (5 x 5 mtx) ...
Performing LU fact. (5 x 5 mtx) - done. Time (sec): 0.0000000
Initializing linear solver - done. Time (sec): 0.0000000
Solving linear system (col. 0) ...
Back solving (5 x 5 mtx) ...
Back solving (5 x 5 mtx) - done. Time (sec): 0.0000000
Solving linear system (col. 0) - done. Time (sec): 0.0000000
Training - done. Time (sec): 0.0000000
___________________________________________________________________________
Evaluation
# eval points. : 100
Predicting ...
Predicting - done. Time (sec): 0.0000000
Prediction time/pt. (sec) : 0.0000000
SurrogateModel class API¶
All surrogate modeling methods implement the following API, though some of the functions in the API are not supported by all methods.
- class smt.surrogate_models.surrogate_model.SurrogateModel(**kwargs)[source]¶
Base class for all surrogate models.
Examples
>>> from smt.surrogate_models import RBF >>> sm = RBF(print_training=False) >>> sm.options['print_prediction'] = False
- Attributes:
- optionsOptionsDictionary
Dictionary of options. Options values can be set on this attribute directly or they can be passed in as keyword arguments during instantiation.
- supportsdict
Dictionary containing information about what this surrogate model supports.
Methods
predict_derivatives(x, kx)Predict the dy_dx derivatives at a set of points.
Predict the derivatives dy_dyt at a set of points.
Predict the output values at a set of points.
predict_variance_derivatives(x, kx)Predict the derivation of the variance at a point
Predict the variances at a set of points.
set_training_derivatives(xt, dyt_dxt, kx[, name])Set training data (derivatives).
set_training_values(xt, yt[, name])Set training data (values).
train()Train the model
update_training_derivatives(dyt_dxt, kx[, name])Update the training data (values) at the previously set input values.
update_training_values(yt[, name])Update the training data (values) at the previously set input values.
- __init__(**kwargs)[source]¶
Constructor where values of options can be passed in.
For the list of options, see the documentation for the surrogate model being used.
- Parameters:
- **kwargsnamed arguments
Set of options that can be optionally set; each option must have been declared.
Examples
>>> from smt.surrogate_models import RBF >>> sm = RBF(print_global=False)
- set_training_values(xt: ndarray, yt: ndarray, name=None) None[source]¶
Set training data (values).
- Parameters:
- xtnp.ndarray[nt, nx] or np.ndarray[nt]
The input values for the nt training points.
- ytnp.ndarray[nt, ny] or np.ndarray[nt]
The output values for the nt training points.
- namestr or None
An optional label for the group of training points being set. This is only used in special situations (e.g., multi-fidelity applications).
- set_training_derivatives(xt: ndarray, dyt_dxt: ndarray, kx: int, name: str | None = None) None[source]¶
Set training data (derivatives).
- Parameters:
- xtnp.ndarray[nt, nx] or np.ndarray[nt]
The input values for the nt training points.
- dyt_dxtnp.ndarray[nt, ny] or np.ndarray[nt]
The derivatives values for the nt training points.
- kxint
0-based index of the derivatives being set.
- namestr or None
An optional label for the group of training points being set. This is only used in special situations (e.g., multi-fidelity applications).
- predict_values(x: ndarray) ndarray[source]¶
Predict the output values at a set of points.
- Parameters:
- xnp.ndarray[nt, nx] or np.ndarray[nt]
Input values for the prediction points.
- Returns:
- ynp.ndarray[nt, ny]
Output values at the prediction points.
- predict_derivatives(x: ndarray, kx: int) ndarray[source]¶
Predict the dy_dx derivatives at a set of points.
- Parameters:
- xnp.ndarray[nt, nx] or np.ndarray[nt]
Input values for the prediction points.
- kxint
The 0-based index of the input variable with respect to which derivatives are desired.
- Returns:
- dy_dxnp.ndarray[nt, ny]
Derivatives.
- predict_output_derivatives(x: ndarray) dict[source]¶
Predict the derivatives dy_dyt at a set of points.
- Parameters:
- xnp.ndarray[nt, nx] or np.ndarray[nt]
Input values for the prediction points.
- Returns:
- dy_dytdict of np.ndarray[nt, nt]
Dictionary of output derivatives. Key is None for derivatives wrt yt and kx for derivatives wrt dyt_dxt.
How to save and load trained surrogate models¶
The SurrogateModel API does not contain any save/load interface. Therefore the user has to handle these operations by him/herself. Below some tips to implement save and load.
For models written in pure Python¶
These operations can be implemented using the pickle module.
Saving the model¶
sm = KRG()
sm.set_training_values(xtrain, ytrain)
sm.train()
filename = "kriging.pkl"
with open(filename, "wb") as f:
pickle.dump(sm, f)
Loading the model¶
sm2 = None
filename = "kriging.pkl"
with open(filename, "rb") as f:
sm2 = pickle.load(f)
For models written in C++ (namely IDW, RBF, RMTB and RMTC)¶
These models can be cached using their data_dir option. Provided the user gives the same training values the model is not retrained but reloaded from cache directory. So by saving the cache directory and the training data, one is able to avoid the training cost and reload the model from cached data.
Saving the model¶
sm = RBF(data_dir="./cache")
sm.set_training_values(xtrain, ytrain)
sm.train()
Loading the model¶
sm2 = RBF(data_dir="./cache")
sm2.set_training_values(xtrain, ytrain) # same training data as above!
sm2.train() # actual training is skipped, cached data model is loaded